
A brief history lesson: pandas read_sql
pandas has over 50 input / output methods and functions (read_excel, read_parquet, read_sas, etc.), and three of those are functions that let you read from a wide variety of databases:
- read_sql
- read_sql_query
- read_sql_table
In this article we’re not discussing the connector libraries different databases have come up with on their own — See, for example, our article on Snowflake’s Python connector. In general, you can either use pandas read_sql, or database-specific connector libraries when those exist.
Why are there three read_sql functions, and how do they differ?
Let’s jump in a time machine back to early 2014. Several of the pandas maintainers (Joris Van den Bossche, Andy Hayden, Damien Garaud, Dan Allan, Jeff Reback, and others) have a problem: The previous year, they’d started incorporating SQLAlchemy into pandas as a way of helping pandas read from multiple database engines, but the result was a few different sql functions that end users could easily confuse. How should they relate to each other? What should the user experience be like?
They decided to create a top-level API called read_sql that, depending on what arguments it’s given, will call read_sql_query or read_sql_table under the hood. If someone knows they want read_sql_query or read_sql_table, they can just call those directly — they’re not strictly helper functions.
So here was the result:
- pandas read_sql — a wrapper function that calls either read_sql_query or read_sql_table
- pandas read_sql_query — a function that lets you pull the results of a SQL query from a database
- pandas read_sql_table — a function that lets you read a table (you can specify whether it’s the whole table or specific columns) from a database
pandas read_sql seems to be built on top of SQLAlchemy, but what is SQLAlchemy?
The SQLAlchemy docs define SQLAlchemy as “a comprehensive set of tools for working with databases and Python,” and much of what pandas read_sql does is take input from users and pass it to SQLAlchemy so that SQLAlchemy can execute commands in the database and return results to pandas.
SQLAlchemy (and thus pandas read_sql) supports several SQL dialects out of the box: Microsoft SQL Server, MySQL / MariaDB, Oracle, PostgreSQL, and SQLite.
To work with other “external dialects” (including Redshift, Snowflake, CockroachDB, etc.), SQLAlchemy will need to call on a suite of related libraries.
SQLAlchemy has two ways of interacting with it:
- Core — The main functionality needed to generate and execute SQL statements, and
- Object-Relational Mapper (ORM) — The system built on Core that lets you take flexibly defined Python objects and map them to database schemas
But with pandas read_sql, you will almost certainly be interacting with the Core side, so don’t worry about the ORM.
When you use pandas read_sql, what is SQLAlchemy doing under the hood?
If a user passes the text of a SQL query to pandas read_sql, then SQLAlchemy just accepts that and executes it without having to translate it into SQL.
But if the user passes over what’s called a SQLAlchemy Selectable (”a SQL construct that represents a collection of rows” — so not a text SQL query, or a pandas statement, but another kind of object that can generate SQL queries), then SQLAlchemy will translate that into a query before executing it in the database and returning the results.
For example, if you decided to pass the selectable:
select(user_table.c.name, user_table.c.fullname)into pandas read_sql, then pandas read_sql would hand that over to SQLAlchemy, and before executing it, SQLAlchemy would translate that into:
SELECT user_account.name, user_account.fullname
FROM user_accountWhat arguments do you need to hand over to pandas read_sql to get it to work?
The only two required parameters when calling pandas read_sql are:
- “sql”: Either the table name you want to pull, or the SQL query you want executed — we discussed this briefly above
- “con”: The database connection
As we mentioned above, if you hand over a table name, pandas read_sql will implicitly call read_sql_table, and if you hand over a SQL query, it will call pandas read_sql_query. (Note that pulling an entire table with pandas read_sql_table leads to the same result as passing the query "SELECT * FROM TABLE" to read_sql_query.) This part is fairly straightforward.
But getting the second required part to work — the connection — can be a little more involved, particularly if you're connecting to an external database, because you often need to find your username, the database name, the password, etc., and make sure they’re all formatted the right way.
In the examples that follow, we use SQLAlchemy to create an in-memory SQLite database and load a table into it, which is much easier than connecting to a database hosted externally. But if you’re looking for an example of how to connect to an external database, here’s one for connecting to Snowflake.
Let’s see pandas read_sql in action!
Before we test out read_sql, we need to have data in a database to work with! We get this by using SQLAlchemy to create an in-memory sqlite engine, and then we load a US government fuel economy dataset that lists emissions test results and other characteristics for many cars by make / model / year, etc., over the past few decades. Ponder Advisor Matt Harrison has a version saved in his Github repo, so we’ll pull from there. And then we use to_sql to store this as a table in our newly created database.
import pandas as pd
from sqlalchemy import create_engine
engine = create_engine('sqlite:///:memory:')
df = pd.read_csv('https://github.com/mattharrison/datasets/raw/master/data/vehicles.csv.zip')
df.to_sql('vehicles', con=engine, if_exists='replace', index=False)Let’s ask how many pre-2010 unique vehicles are in this dataset. (”id” is the unique identifier.)
Using a pandas dataframe
First we’ll do this with a dataframe so we have a source of truth to compare to.
df.loc[df['year']<2010].id.nunique()28636Using pandas read_sql + a SQL query
Now let’s do this with pandas read_sql, by passing in a SQL query:
pd.read_sql('SELECT COUNT(id) FROM vehicles WHERE year < 2010', engine) COUNT(id)
0 28636Here, under the hood, read_sql called on read_sql_query, and passed the text of the query to that.
Using pandas read_sql + a SQLAlchemy Selectable
And finally, let’s get a little more advanced, and instead pass in a SQLAlchemy Selectable. To do this, we have to import a few additional functions from SQLAlchemy:
from sqlalchemy import select, MetaData, Table, funcWe’ve already created a vehicles table in our in-memory database, but Python wouldn’t recognize that database if we just typed in something like type(vehicles) — It’s not saved as a Python object yet. So we have to create a SQLAlchemy table object that Python can recognize and work with.
# We create an empty MetaData object merely because Table requires one
metadata = MetaData()
table = Table(
"vehicles",
metadata,
autoload_with=engine
)Finally, we use the SQLAlchemy syntax to generate our Selectable. We usefunc.count()ontable.c.idto count the number of ids, and then we wrap that withselect(). Finally, we add.where()on the end to subset on only those rows where the year is less than 2010.
statement = select(func.count(table.c.id)).where(table.c.year < 2010)
pd.read_sql(statement, con=engine.connect()) count_1
0 28636And we get the same answer once again. Pretty cool!
What if pulling data from my database into memory is slow, costly, or just not possible?
This is where Ponder comes in — Sometimes tables are too large to pull into memory as pandas dataframes, or even if it’s technically feasible, pulling them in is slow or costly or poses a security risk.
Ponder has reimplemented the internals of the pandas API so that you don’t need to pull your data out of your warehouse, and your pandas commands instead get executed as SQL directly in your warehouse.
So Ponder’s implementation of pandas read_sql will create a pointer to your table in your data warehouse instead of pulling the table out of your warehouse and into memory.
This can lead to massive workflow speedups, like the 2-HOUR to 2-MINUTE speedup we documented in an earlier post:
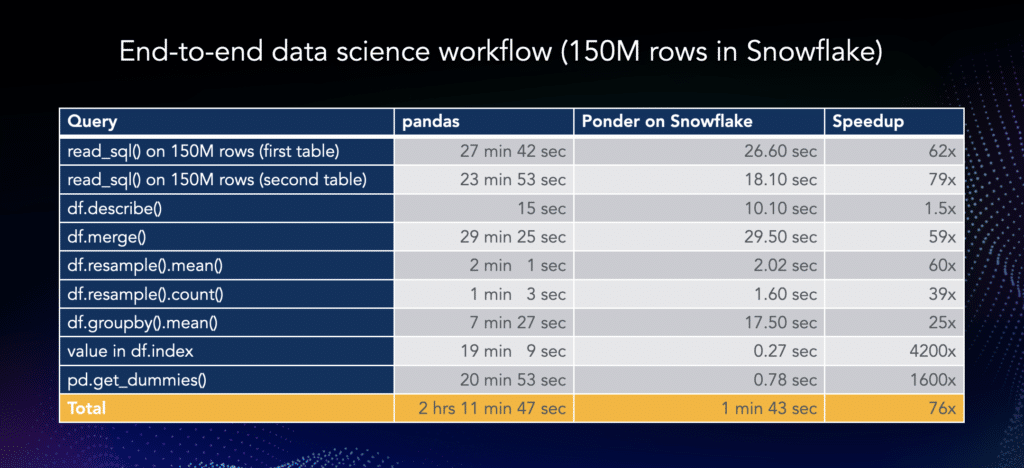
If you’d like to see pandas read_sql at its best, try Ponder today!
Try Ponder Today
Start running your Python data science workflows in your database within minutes!




