
Summary
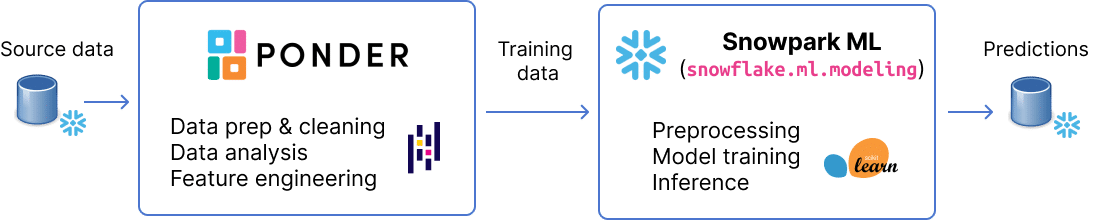
tl;dr: In this post, we walk through an end-to-end machine learning workflow to show how you can use Ponder and Snowpark ML to analyze electronic health records directly in your data warehouse.
We build off of our earlier blogpost to demonstrate how you can use Snowflake’s exciting new Snowpark ML modeling API — in public preview as of June’s Snowflake Summit — to perform ML in your database, rather than having to pull your data out of Snowflake to use sklearn in-memory.
You can download the notebook associated with this post here.
The MIMIC-III Clinical Dataset¶
In this blog post, we will be looking at the MIMIC-III demo dataset. The MIMIC-III Clinical Database contains deidentified health-related data of patients who stayed in an intensive care unit (ICU) at the Beth Israel Deaconess Medical Center in Boston. The demo dataset contains records for 100 patients across three tables PATIENTS, ICUSTAYS, and ADMISSIONS. You can download the datasets here or use our dataset upload script to load it to Snowflake.
What is Ponder?¶
Ponder lets you run pandas directly in your data warehouse. Data teams can interact with their data through their familiar pandas-native experience, while enjoying the scalability and security benefits that come with a modern cloud data warehouses. You can learn more about Ponder in our recent blogpost and sign up here to try out Ponder today.
Try Ponder Today
Start running your Python data science workflows in your database within minutes!
import os; os.chdir("..")
import credential
import ponder
ponder.init()
credential.params["database"] = "MIMIC3"
import snowflake.connector
# Create a Ponder Snowflake Connections object
snowflake_con = snowflake.connector.connect(
user=credential.params["user"],
password=credential.params["password"],
account=credential.params["account"],
role=credential.params["role"],
database=credential.params["database"],
schema=credential.params["schema"],
warehouse=credential.params["warehouse"]
)
import modin.pandas as pd
icu = pd.read_sql("ICUSTAYS",con=snowflake_con)
patients = pd.read_sql("PATIENTS",con=snowflake_con)
df = patients.merge(icu,on="subject_id")
df = df.drop(list(df.filter(regex="row_id")),axis=1)
Exploratory Data Analysis + Preprocessing
Dataset Overview
df.describe()
| subject_id | expire_flag | hadm_id | icustay_id | first_wardid | last_wardid | los | |
|---|---|---|---|---|---|---|---|
| count | 136.000000 | 136.0 | 136.000000 | 136.000000 | 136.000000 | 136.000000 | 136.000000 |
| mean | 28263.485294 | 1.0 | 153259.566176 | 250980.470588 | 34.470588 | 34.022059 | 4.452457 |
| std | 16008.281510 | 0.0 | 28054.220280 | 28455.125832 | 18.876182 | 19.280951 | 6.196828 |
| min | 10006.000000 | 1.0 | 100375.000000 | 201006.000000 | 7.000000 | 7.000000 | 0.105900 |
| 25% | 10089.750000 | 1.0 | 129028.000000 | 224359.250000 | 14.750000 | 14.000000 | 1.233525 |
| 50% | 40307.000000 | 1.0 | 157724.000000 | 250055.000000 | 33.000000 | 33.000000 | 2.111450 |
| 75% | 42207.000000 | 1.0 | 174896.500000 | 277279.250000 | 52.000000 | 52.000000 | 4.329050 |
| max | 44228.000000 | 1.0 | 199395.000000 | 298685.000000 | 57.000000 | 57.000000 | 35.406500 |
df.info()
<class 'modin.pandas.dataframe.DataFrame'> RangeIndex: 136 entries, 0 to 135 Data columns (total 17 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 subject_id 136 non-null int32 1 gender 136 non-null object 2 dob 136 non-null object 3 dod 136 non-null object 4 dod_hosp 88 non-null object 5 dod_ssn 108 non-null object 6 expire_flag 136 non-null int8 7 hadm_id 136 non-null int32 8 icustay_id 136 non-null int32 9 dbsource 136 non-null object 10 first_careunit 136 non-null object 11 last_careunit 136 non-null object 12 first_wardid 136 non-null int8 13 last_wardid 136 non-null int8 14 intime 136 non-null object 15 outtime 136 non-null object 16 los 136 non-null float64 dtypes: float64(1), int32(3), int8(3), object(10) memory usage: 0.0+ bytes
df["intime"] = pd.to_datetime(df["intime"])
df["outtime"] = pd.to_datetime(df["outtime"])
df["dob"] = pd.to_datetime(df["dob"])
df["length_of_stay"] = (df["outtime"]-df["intime"])/pd.Timedelta('1 hour')
df["age"] = df["intime"].dt.year-df["dob"].dt.year
df = df[df["age"]<100]
df.plot("age","length_of_stay",kind="scatter")
<Axes: xlabel='age', ylabel='length_of_stay'>
admissions = pd.read_sql("ADMISSIONS",con=snowflake_con)
df = df.merge(admissions,on=["hadm_id","subject_id"])
df["pre_icu_length_of_stay"]= (df["intime"]-df["admittime"])/pd.Timedelta('1 day')
df["pre_icu_length_of_stay"].hist()
<Axes: >
print(f"Percentage of ICU admissions within 1 day: \
{len(df[df['pre_icu_length_of_stay']<1])/len(df)*100:.2f}%")
Percentage of ICU admissions within 1 day: 81.10%
You can find the SQL that performs a similar query on BigQuery in this tutorial.
Working with Text: Parsing through clinical diagnosis¶
df.diagnosis
0 SEPSIS
1 HEPATITIS B
2 SEPSIS
3 HUMERAL FRACTURE
4 ALCOHOLIC HEPATITIS
...
122 SHORTNESS OF BREATH
123 PERICARDIAL EFFUSION
124 ACUTE RESPIRATORY DISTRESS SYNDROME;ACUTE RENA...
125 BRADYCARDIA
126 CHOLANGITIS
Name: diagnosis, Length: 127, dtype: object
df.diagnosis = df.diagnosis.str.replace(";"," ").str.strip()
all_diagnosis_str = df.diagnosis.str.cat(sep=" ")
subs = {"\\":" ", "-":"", "/":"", "?":""}
for s in subs.keys():
all_diagnosis_str = all_diagnosis_str.replace(s, subs[s])
all_diagnosis = all_diagnosis_str.split(" ")
import collections
c = collections.Counter(all_diagnosis)
c.most_common(5)
[('FAILURE', 12), ('SEPSIS', 11), ('PNEUMONIA', 9), ('BLEED', 8), ('ACUTE', 7)]
top_5_keyword = sorted(c, key=c.get, reverse=True)[:5]
print(f"Top 5 most common diagnostic terms are: {top_5_keyword}")
Top 5 most common diagnostic terms are: ['FAILURE', 'SEPSIS', 'PNEUMONIA', 'BLEED', 'ACUTE']
for keyword in top_5_keyword:
df[keyword]=df['diagnosis'].str.contains(keyword)
df["hospital_expire_flag"].value_counts()
hospital_expire_flag 0 85 1 42 Name: count, dtype: int64
X = df[top_5_keyword].astype(int)
y = df['hospital_expire_flag']
dataset = pd.concat([X,y],axis=1)
dataset = dataset.rename(columns={"hospital_expire_flag":"TARGET"})
dataset
| FAILURE | SEPSIS | PNEUMONIA | BLEED | ACUTE | TARGET | |
|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| 2 | 0 | 1 | 0 | 0 | 0 | 1 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 1 |
| … | … | … | … | … | … | … |
| 122 | 0 | 0 | 0 | 0 | 0 | 0 |
| 123 | 0 | 0 | 0 | 0 | 0 | 0 |
| 124 | 1 | 0 | 0 | 0 | 1 | 0 |
| 125 | 0 | 0 | 0 | 0 | 0 | 0 |
| 126 | 0 | 0 | 0 | 0 | 0 | 0 |
dataset.to_sql("ICU_MORTALITY",con=snowflake_con, index = None, if_exists="replace")
Machine Learning with Snowpark ML: Mortality prediction of ICU Patients¶
What is Snowpark ML Modeling?¶
Snowpark ML Modeling gives users a Python interface for preprocessing and training models. Similar to how Ponder runs all your pandas operations directly in Snowflake, Snowpark ML modeling runs all your Python ML operations directly in Snowflake. Unlike Ponder, this is done by registering your Python models as user-defined function to be executed on the database. This means that data doesn’t leave your warehouse and you inherit the scale, performance, and security you get with Snowflake. (Note: While Snowpark ML supports preprocessing procedures, since we already performed all the preprocessing in pandas using Ponder, we only use the modeling APIs here)from snowflake.snowpark import Session
session = Session.builder.configs(credential.params).create()
dataset = session.table("ICU_MORTALITY")
dataset.show()
--------------------------------------------------------------------- |"FAILURE" |"SEPSIS" |"PNEUMONIA" |"BLEED" |"ACUTE" |"TARGET" | --------------------------------------------------------------------- |0 |1 |0 |0 |0 |0 | |0 |0 |0 |0 |0 |1 | |0 |1 |0 |0 |0 |1 | |0 |0 |0 |0 |0 |0 | |0 |0 |0 |0 |0 |1 | |0 |0 |0 |0 |0 |0 | |0 |0 |0 |0 |0 |0 | |0 |0 |0 |0 |0 |0 | |0 |0 |0 |0 |0 |0 | |0 |0 |0 |0 |0 |0 | ---------------------------------------------------------------------
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size= 0.1,random_state=0)
from sklearn.naive_bayes import GaussianNB
model = GaussianNB()
model.fit(X_train,y_train)
predictions = model.predict(X_test)
# Split the data into train and test sets
train_df, test_df = dataset.random_split(weights=[0.88, 0.12], seed=0)
print (f"Training size: {train_df.count()} , Test size: {test_df.count()}")
Training size: 114 , Test size: 13
Now, we are ready to build our model. The Snowpark ML API is largely inspired by three popular Python ML libraries: scikit-learn, xgboost, and lightgbm. In this blogpost, we will only be using the scikit-learn APIs. We will show the corresponding code for performing the same operation in scikit-learn. You may note some key differences, in particular, in the model initialization step, Snowpark requires that you specify the input and label columns upfront (i.e., input being the features and label being the target variable you are trying to predict). In scikit-learn, you will notice that this happens on the fit step rather than in the model definition step. You can read more about the API differences here.
from snowflake.ml.modeling.naive_bayes import GaussianNB
model = GaussianNB(input_cols=['FAILURE', 'SEPSIS', 'PNEUMONIA', 'BLEED', 'ACUTE'],label_cols="TARGET")
We fit a basic Naive Bayes classifier and evaluate the model predictions.
model = model.fit(train_df)
predictions = model.predict(test_df)
predictions.show()
--------------------------------------------------------------------------------------- |"TARGET" |"SEPSIS" |"PNEUMONIA" |"ACUTE" |"BLEED" |"FAILURE" |"OUTPUT_TARGET" | --------------------------------------------------------------------------------------- |1 |0 |0 |0 |1 |0 |1 | |0 |0 |0 |0 |0 |0 |1 | |1 |0 |0 |0 |0 |0 |1 | |1 |0 |0 |0 |0 |0 |1 | |0 |1 |1 |0 |0 |0 |0 | |0 |0 |0 |0 |0 |0 |1 | |0 |0 |1 |0 |0 |0 |0 | |0 |0 |0 |0 |0 |0 |1 | |0 |1 |0 |0 |0 |0 |0 | |1 |0 |0 |0 |0 |0 |1 | ---------------------------------------------------------------------------------------
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
cm = confusion_matrix(y_test, predictions)
disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=model.classes_)
disp.plot()
from snowflake.ml.modeling.metrics import confusion_matrix
cm = confusion_matrix(df= predictions, y_pred_col_name= "OUTPUT_TARGET", y_true_col_name="TARGET")
cm
array([[3., 6.],
[0., 4.]])
from sklearn.metrics import ConfusionMatrixDisplay
disp = ConfusionMatrixDisplay(confusion_matrix=cm)
disp.plot()
<sklearn.metrics._plot.confusion_matrix.ConfusionMatrixDisplay at 0x14b10f8b0>
from sklearn.metrics import accuracy_score
score = accuracy_score(y_test, predictions)
print(f'Accuracy of the binary classifier = {score:0.2f}')
from snowflake.ml.modeling.metrics import accuracy_score
score = accuracy_score(df= predictions, y_pred_col_names= "OUTPUT_TARGET", y_true_col_names="TARGET")
print(f'Accuracy of the binary classifier = {score:0.2f}')
Accuracy of the binary classifier = 0.54
Why Ponder + Snowpark ML?
- browse high-level summary and overview of the dataset,
- discover patterns and insights based on visualizations and basic statistics,
- perform date time operations to compute patient’s length of stay,
- develop features based on clinician free-text diagnosis.
Looking to try Ponder for your project? Sign up here to get started in using Ponder!
Try Ponder Today
Start running your Python data science workflows in your database within minutes!




